Preparation
Once your Soteria request form has been reviewed and approved, you will receive an email with the subject UA Soteria Access Request Approved. This email will contain the next steps to take which are detailed below:
Complete Required Training in Edge Learning
The CRRSP team will register you for the required trainings listed below (courses can also be found here: https://uaccess.arizona.edu):
- HIPAA Essentials
- Information Security: Insider Threat Awareness
- Information Security Awareness Certification
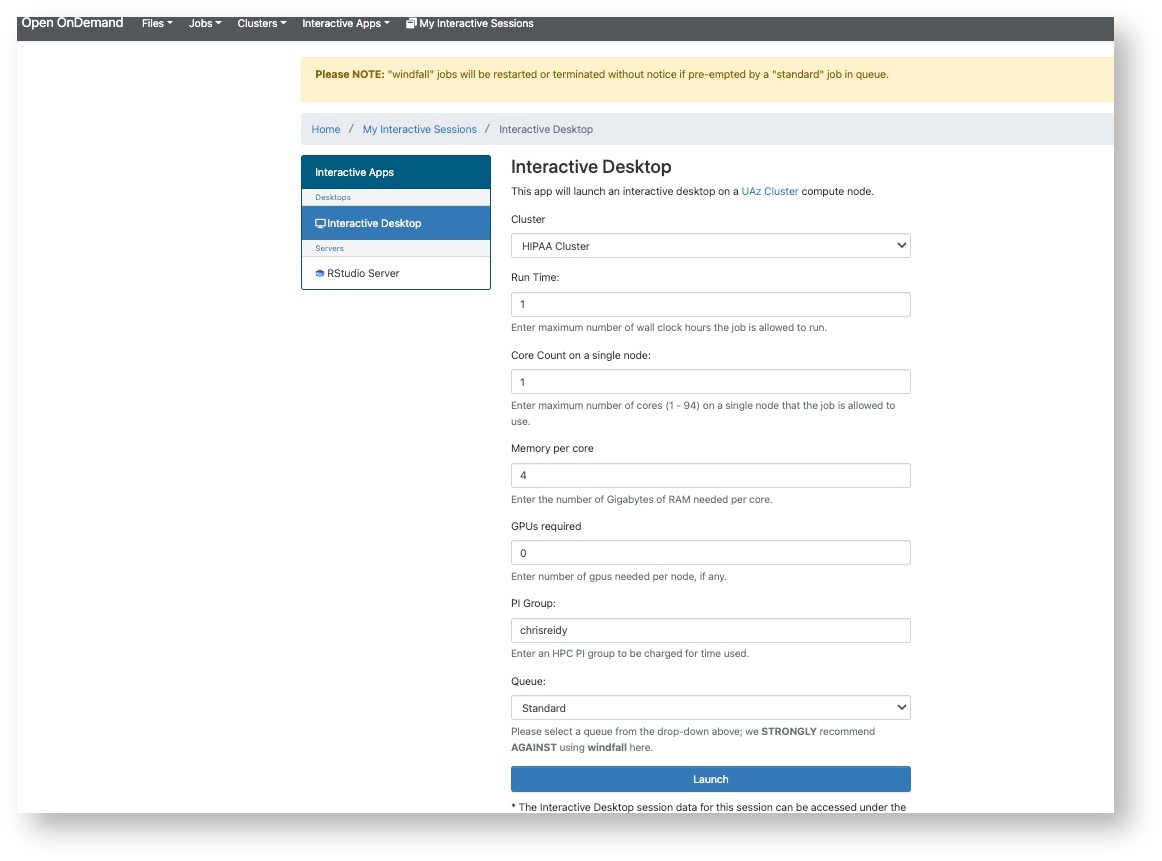
Assignment to the Soteria VPN
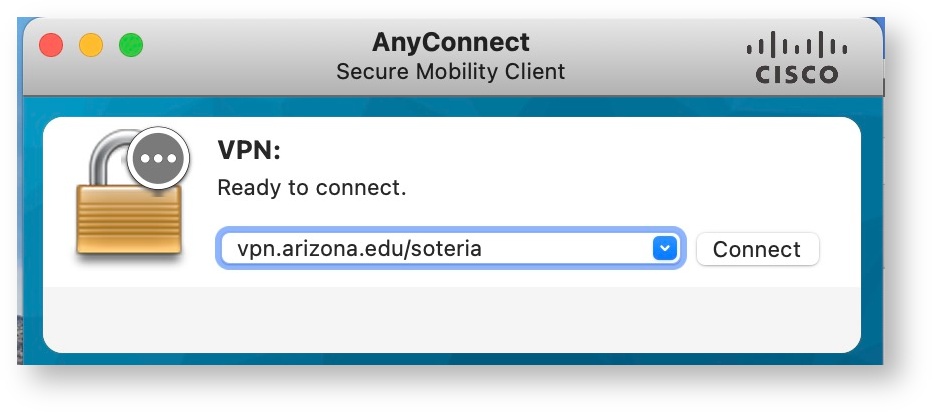
Once you have completed your required training, the CRRSP team will notify you via email when you have been assigned access to the Soteria VPN. This VPN is an important part of our HIPAA compliance and differentiates Soteria usage from the standard HPC clusters. Soteria access cannot be established when not connect to the VPN. For VPN access, use: vpn.arizona.edu/soteria.
Additional Instructions
The computer you will use to access Soteria services must meet the following requirements:
- The Operating System and applications must be updated with the latest patches
- You must have a strong password to log into the computer (at least 8 characters and a mix of character types).
- This must not be a shared computer with other users
- Up to date anti-virus software
Resources
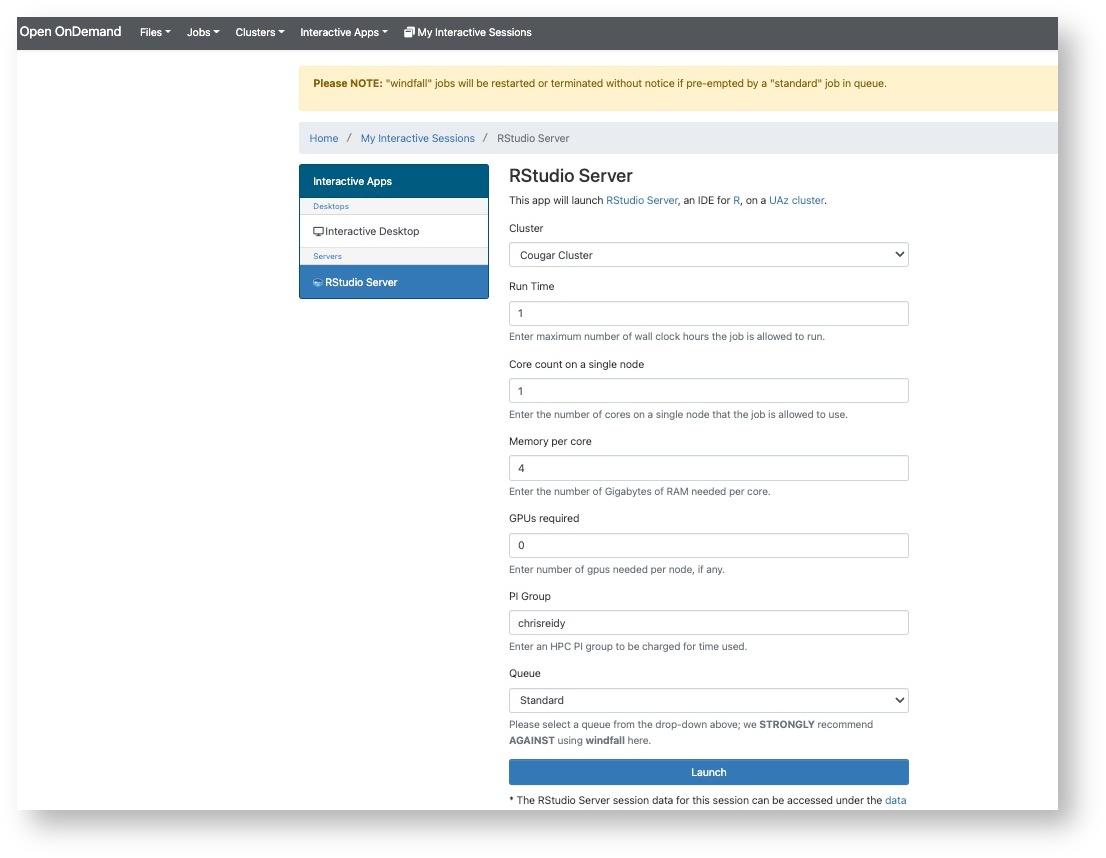
The cluster has four standard compute nodes. Each has 94 cores and 512GB memory available. The two GPU nodes have the same resources but there are also four V100 GPU's in each. You can use the regular parts of the documentation to learn how to use slurm with these nodes.
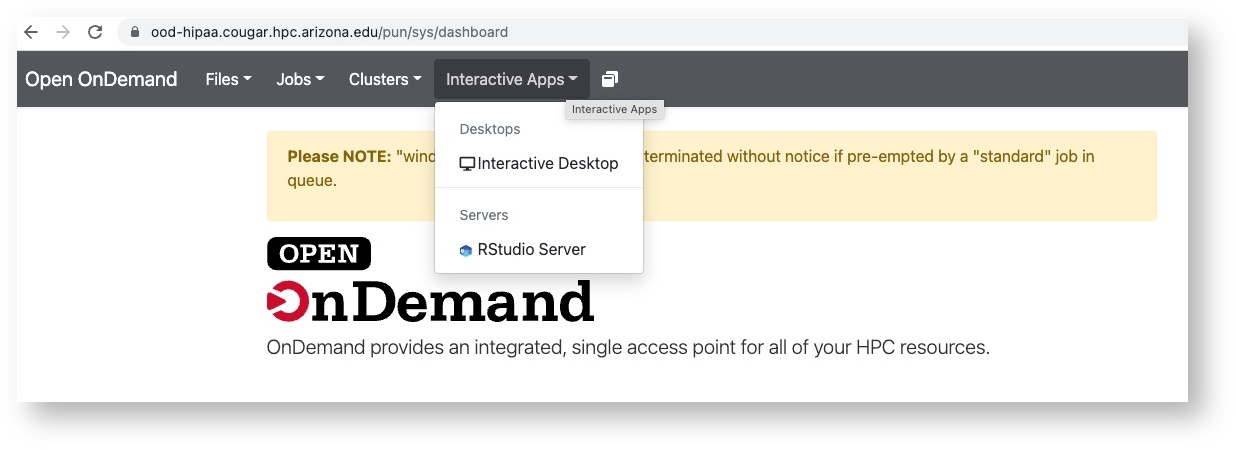
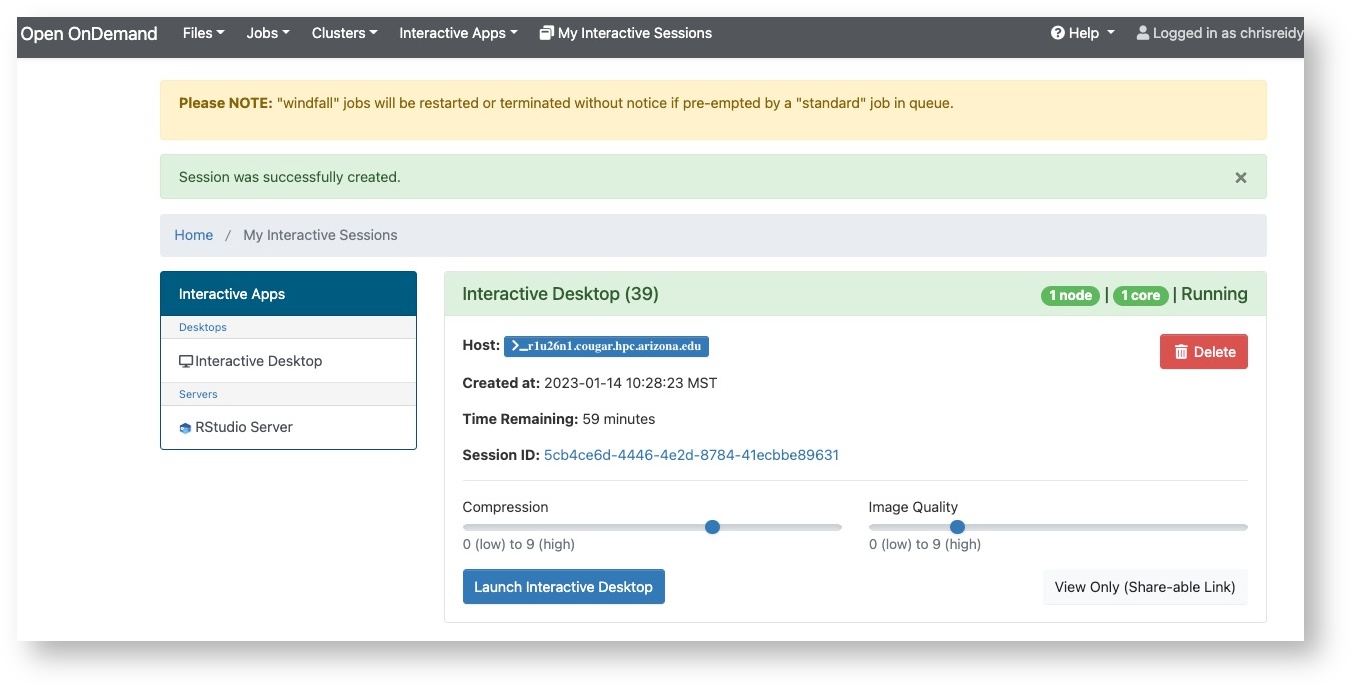
In case you are interested, these nodes are named by their physical location and you will see this if you are connected to an interactive session. And with OnDemand (OOD) your connection will show the node hostname.
| Standard Nodes | GPU Nodes |
|---|---|
| r1u26n1,r1u27n1,r1u28n1,r1u29n1 | r1u30n1,r1u32n1 |
For the purpose of this early testing, the allocations of time and space will be similar to HPC.
The time allocation will be 100,000 hours.
Your account will come with space in /home and /groups where you can put your data. Currently those directories are not subject to a quota limit.
System Access
You must be connected to the Soteria VPN (see section above) to access the system.
Transferring and Accessing Data
Globus
Globus can be used for moving data in and out of the Soteria environment. For more information on using Globus: https://uarizona.atlassian.net/wiki/display/UAHPC/Transferring+Data#TransferringData-GridFTP/Globus
Soteria's endpoint is: UA HPC HIPAA Filesystems
File Paths
Your files can be accessed on the filexfer nodes:
- /hipaa/groups/<pi_netid>
- /hipaa/home/uxx/<your_netid>
When connected to a Soteria login/compute node, you can find these under:
- /groups/<pi_netid>
- /home/uxx/<your_netid>
Getting Help
We use ServiceNow and can be reached with a support ticket.